- Investigation of Secondary Structure and Thermodynamic Properties of Antitoxin PrlF and Toxin YhaV in Escherichia coli
*4D Convergence Technology Institute (National Key Technology Institute in University),
Korea National University of Transportation, Jungpyeog, Chung-Buk 27909, Korea
**Department of Chemical & Biological Engineering, Korea National University of Transportation,
Chungju, Chung-Buk 27469, Korea- 대장균 항독소 PrlF와 독소 YhaV의 이차구조 및 열안정 조사
*국립한국교통대학교 4D 융합기술연구소, **국립한국교통대학교 화공생물공학과
Reproduction, stored in a retrieval system, or transmitted in any form of any part of this publication is permitted only by written permission from the Polymer Society of Korea.
Toxin-antitoxin (TA) systems are key regulators of bacterial growth and survival under harsh environmental conditions. PrlF-YhaV is type II TA system in Escherichia coli. However, the structures of antitoxin PrlF and toxin YhaV in the PrlF-YhaV TA system remain poorly understood. Here, we explored the secondary structure and thermal stability of antitoxin PrlF and toxin YhaV by circular dichroism (CD) spectroscopy. Analysis of the secondary structure, conducted at wavelengths between 190 to 260 nm at 25 °C, revealed that antitoxin PrlF comprises 41.2% helical and 29.3% β-sheet structural conformations. In contrast, toxin YhaV primarily assumes a helical conformation, making up 85.7% of its structure. Thermal stability analysis demonstrated that the melting points of antitoxin PrlF and toxin YhaV are 50 °C and 75 °C, respectively, as observed at 200 or 222 nm. Additionally, antitoxin PrlF exhibited reversible structural formations, while the toxin YhaV did not regain the structural reversibility under refolding state of 90 to 25 °C. Our examination of secondary structure components suggests that the β-sheet of antitoxin PrlF and α-helix of toxin YhaV play key roles in their respective structural configurations and thermal stabilities. These findings clarify the distinct physiological characteristics of these two proteins, emphasizing their differing structures and thermal stabilities across various temperature conditions in TA loci.
독소-항독소 시스템은 박테리아가 가혹한 환경 조건에서 생존하고 성장을 조절하는 데 핵심적인 역할을 하며, PrlF-YhaV는 대장균에서 발견된 II형 독소-항독소 시스템이다. 그러나 PrlF 항독소와 YhaV 독소의 구조 및 열 안정성 연구관련 보고된 바 없으며, 본 연구에서는 원형 이색성 분광법을 이용하여 PrlF 항독소와 YhaV 독소의 이차 구조 및 열적 안정성을 조사하였다. 25°C에서 190~260 nm 파장 범위의 스펙트럼 분석 결과, PrlF 항독소는 41.2%의 α-나선 구조와 29.3%의 β-병풍 구조를 포함하고 있음이 확인된 반면, YhaV 독소는 85.7%가 α-나선 구조로 이루어져 있어 주로 나선형 구조를 갖는 것으로 확인되었다. 열 안정성 구조 분석 결과 PrlF 항독소와 YhaV 독소의 융해 온도가 각각 50 °C와 75 °C임이 200 nm 또는 222 nm에서 관찰되었으며, PrlF 항독소는 열변성 후 90 °C에서 25°C로 냉각 시 구조가 회복되는 반면, YhaV 독소는 구조적 가역성을 보이지 않았다. 이차 구조적 특성에 대한 분석을 종합해볼 때, PrlF 항독소의 β-병풍 구조와 YhaV 독소의 α-나선 구조가 각각의 단백질 구조 구성 및 열적 안정성에 중요한 역할을 하는 것으로 판단되며, 이러한 결과는 두 단백질이 독소-항독소 유전자 자리에서 서로 다른 구조적 특징과 열 안정성을 가지고 있음을 명확히 하며, 이들의 생리학적 차이를 규명하는 데 중요한 정보를 제공한다.
This study identifies the structural features that confer thermal stability to the PrlF antitoxin and its cognate YhaV toxin. It shows that PrlF exhibits structural reversibility, whereas YhaV does not regain its structure under refolding conditions. These findings propose a novel mechanism for the YhaV–PrlF toxin–antitoxin system under high-temperature conditions.

Keywords: toxin-antitoxin, PrlF, YhaV, PrlF-YhaV, secondary structure.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (Grant No. RS-2021-NR060137). Additional support was provided by the NRF, funded by the Ministry of Science and ICT (MSIT) of the Korean government (Grant Nos. RS-2024-00405287, 2021R1A2C2095113 and RS-2025-02217286).
The authors declare no potential conflict of interest.
Toxin-antitoxin (TA) systems are a prominent group of proteins or RNAs persistent across various bacterial genera that codes for a stable toxin and a labile antitoxin.1 TA systems are broadly classified into 8 categories depending on the neutralizing characteristics of the antitoxin for the respective toxin.2 PrlF-YhaV is a type II TA system identified in Escherichia coli, where the operon codes for PrlF antitoxin similar to AbrB transcription factor and YhaV toxin which is a RelE superfamily toxin.3 The intrinsic toxin activity is up-regulated under unfavourable conditions and are suppressed by their cognate antitoxins under normal conditions.4,5 For example, TA systems are triggered under nutritional stress, antibiotic exposure, oxidative stress, high temperature, and extracellular death factors that eventually leads to TA system induced programmed cell death due to its toxic nature.5 There are limited studies on the specific unfavourable conditions where PrlF-YhaV TA system are activated. However, this system has been predicted to be activated under antibiotic exposure and elevated temperature.6
The toxin’s mode of action can depend upon their ability to bind to DNA, RNA or protein motifs and inhibit cell growth through various mechanism.6-8 The PrlF-YhaV TA system is a ribosome-dependent mRNA interferase TA system and the toxin acts as a bacteriostatic agent under environmental stress conditions.9 There are other TA systems which belong to the similar category, such as RelBE, YoeB-YefM, HipA-HipB, and YafO-YafN.10 Among them, the RelBE TA system has been well characterized based on their physiology, structure and function.11-13 The structural analysis of RelE has shown the endoribonuclease active site being located in the C-terminus which is important for determining their function.14
The previous studies from our group has characterized the ribosome dependent mRNA interferase activity of YhaV toxin and determined that the C-terminus region is important for such activity.9,15 Additionally, it is important to understand the secondary structure and refolding mechanism of proteins which pertains to the understanding regarding the proper functioning of the respective proteins under varied environmental conditions.16-17 Hence, this study characterized the physiological functioning of the PrlF-YhaV TA system of Escherichia coli in vivo and their secondary structure in vitro at different temperature regimes. This study was also conducted to evaluate the refolding mechanism of both the toxin and the antitoxin proteins when they are subjected to elevated temperatures for determining their mode of action under high temperature. In this study, we considered evaluating the in vitro secondary and refolding mechanism of PrlF and YhaV proteins, as there are limited reports on the characterization of these particular proteins.
Construction of PrlF, YhaV, and PrlF-YhaV Expression Systems. The desired operons, viz., prlF (antitoxin), yhaV (toxin) and the complex prlF-yhaV were amplified with conventional polymerase chain reaction (PCR) from the genomic DNA of E. coli BW 25113 and cloned into pBAD24, pET28a and pET21a expression vectors.9 The successful cloning of the operons in the expression vectors were assessed by transforming them into E. coli DH5αΔprlFyhaV and growing the transformants in luria-bertini (LB) agar plates supplemented with appropriate antibiotics (100 mg/ml ampicillin for pBAD24 and pET21a; 50 mg/mL kanamycin for pET28a).
Assessment of Viability of Bacterial Cells. The bacterial cell viability was determined by inoculating 1/100th volume of overnight grown culture in M9-glycerol broth supplemented with 100 μg mL-1 ampicillin and incubating at 37 ℃ in a shaker incubator. The absorbance was monitored at 600 nm using a UV-Vis spectrophotometer and 0.2% arabinose was added when OD600 reached 0.4 and the bacterial suspension was allowed to grow for 3 h at 37 ℃in a shaker incubator. The bacterial cell suspension was washed and subsequently diluted to 1 × 106 CFU mL with cold phosphate buffer saline (PBS). The diluted samples were stained with 1 mg/mL propidium iodide (550825, BD Biosciences, San Diego, CA, USA) following the described previously.18 The cell viability was recorded on a flow cytometer (BD FACS calibur, BD Biosciences).
Expression and Purification of PrlF Antitoxin and YhaV Toxin. The pET28a-prlF and pET21-prlF-yhaV were transformed into E. coli BL21 (DE3) ΔprlFyhaV for the expression and purification of PrlF antitoxin and YhaV toxin, respectively. The respective transformed cells were grown in LB broth separately in flasks until OD600 reached 0.6 at 37℃, and 1 mM isopropyl-β-D-1-thiogalactoside was added to the cell suspension. The induced cells were incubated at 18 ℃ for 8 h in a shaker incubator. To purify the PrlF antitoxin, the respective induced cells were harvested by centrifugation 4000 × g for 30 min at 40℃, and the harvested cell pellets were resuspended in buffer A 50 mM sodium phosphate buffer, pH 7.8). After cell disruption by French cell press (Vison Scientific Co., LTD, Republic of Korea), the soluble supernatant was prepared by by centrifugation 4000 × g for 30 min at 4℃, and N-terminally histidine (Hisx6)-tagged PrlF was purified using Ni-NTA agarose (Qiagen, Germany) following the manufacturer’s protocol. Purification sample of YhaV toxin was prepared using same induction conditions from pET21-prlF-yhaV. Briefly, the YhaV toxin was purified by solid-phase refolding method with 8 M guanidine. The soluble supernatant of PrlF-YhaV was loaded into packed Ni-NTA agarose resin open column and was bound. It was directly denatured by buffer in 50 mM sodium phosphate pH 7.8 with 8 M guanidine-HCl and was directly refolded by buffer in 50 mM sodium phosphate pH 7.8 on column. Finally, the C- terminally histidine (Hisx6) refolded YhaV toxin was purified using Ni-NTA agarose (Qiagen, Germany) following the manufacturer’s protocol. Refolding method was modified from previous our purification method.19 All the purified proteins were dialyzed by 20 mM Tris-HCl buffer (pH 7.8), respectively.
Determination of Molecular Weights by MALDI-TOF. The molecular weights of the purified PrlF and YhaV were determined by MALDI-TOF mass spectrometer (Bruker Daltonics, Germany) following a method described previously.20 The purified proteins of PrlF and YhaV were prepared by mixing equal volumes of protein solution (NagA) with sinapinic acid (10 mg/mL in 50% ACN/0.1% TFA). 1 μL of each mixture was spotted onto a MALDI plate, after dried in room temperature, it was analyzed by MALDI-TOF mass spectrometer.
Determination of Secondary Structure by Circular Dichroism (CD) Spectroscopy. The secondary structure of purified his-tagged PrlF antitoxin (0.25 mg/mL) and YhaV toxin (0.25 mg/mL) were determined using Far-UV by Chirascan CD spectrometer (Applied Photophysics, UK). The CD spectra of proteins were recorded over a wavelength range of 190 to 260 nm using a 0.5 mm path length from 25 to 90 ℃ and 90 to 25 ℃of increasing or decreasing step by step 5 ℃ interval. All experiments were conducted in 20 mM Tris-HCl buffer (pH 7.8). The melting points were determined by 0.2 ℃ gradient increase in temperature from 25 to 90 ℃. Each CD spectrums were calculated by subtracted the background CD signal the buffer. The secondary structure was finally determined by CDNN program (Version 2, Applied Photophysics, UK).21
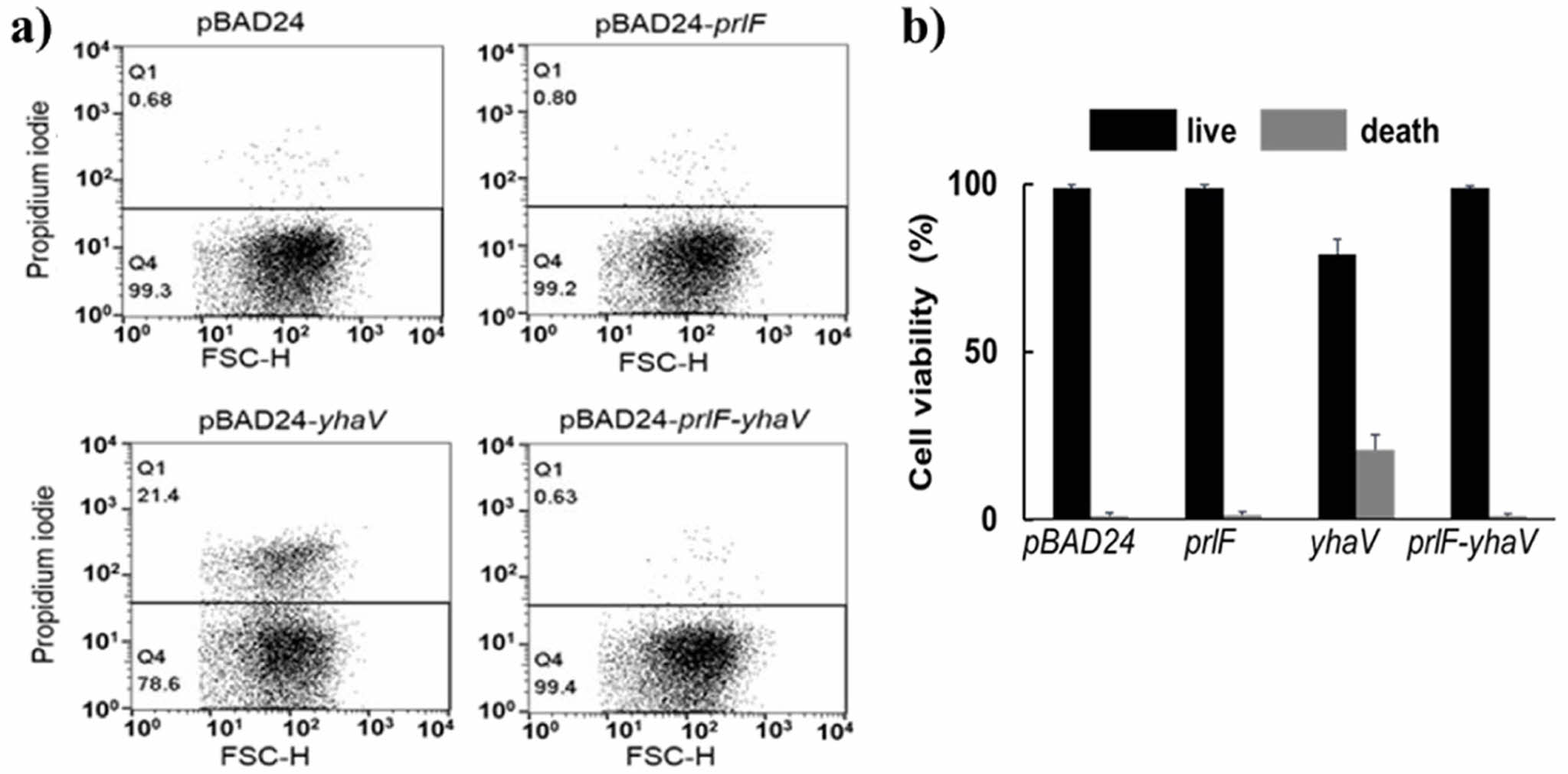
PrlF Antitoxin Neutralized the Toxic Effect of YhaV In Vivo. PrlF-YhaV is a well-known TA system found in E. coli and its function has been well characterized previously.3,9,15 To confirm the toxic effect of YhaV and the toxin neutralization efficacy of PrlF in vivo, we evaluated the viability of E. coli cells expressing YhaV and PrlF-YhaV. The E. coli cells transformed with pBAD24-yhaV recorded about 78.6% cell viability, whereas the cells harbouring empty vector (pBAD), pBAD-prlF and pBAD-prlF-yhaV showed over 99% cell viability, as shown in Figure 1. The previous reports have suggested that the YhaV toxin retards the growth of cells through bactericidal mechanism, which is as typical feature of various other bacterial TA systems in vivo. However, our results suggest that the YhaV toxin acts as a bacteriostatic agent resulting in growth arrest of E. coli cells. As PrlF-YhaV belongs to the type II TA system, their bacteriostatic characteristics can be attributed to translation rescue, ribosome hibernation, increase in cell wall thickness and reduced cell division that might lead to bacterial dormancy for suspending unfavourable conditions. However, extensive research is required to identify the exact mechanism by which PrlF-YhaV inhibits bacterial growth.
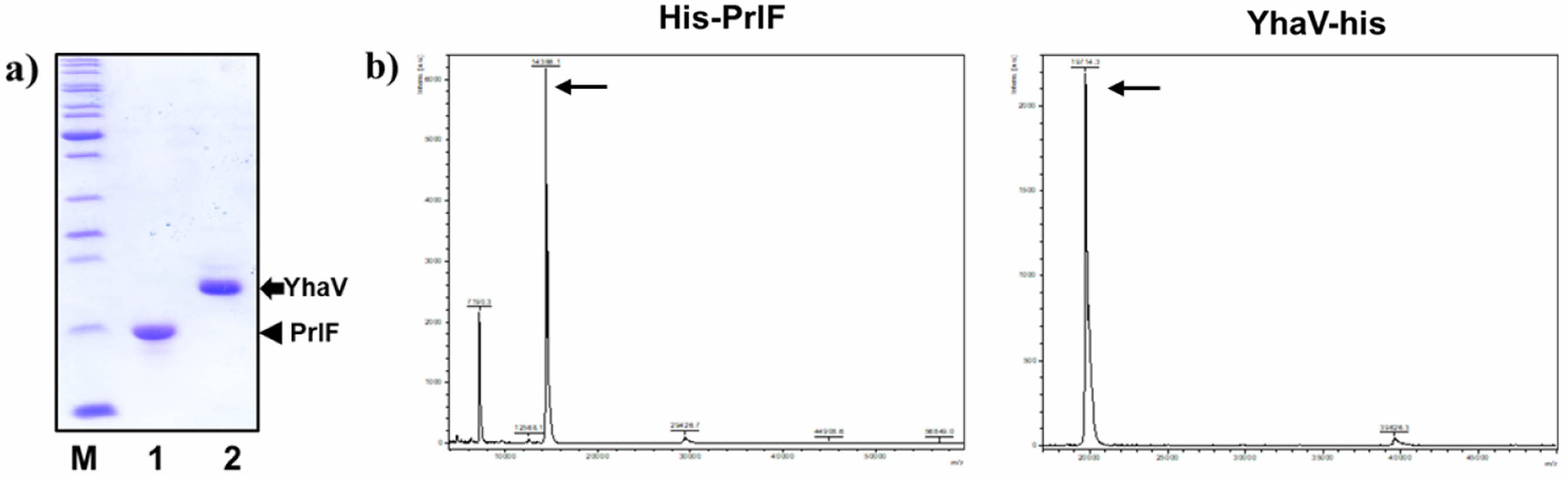
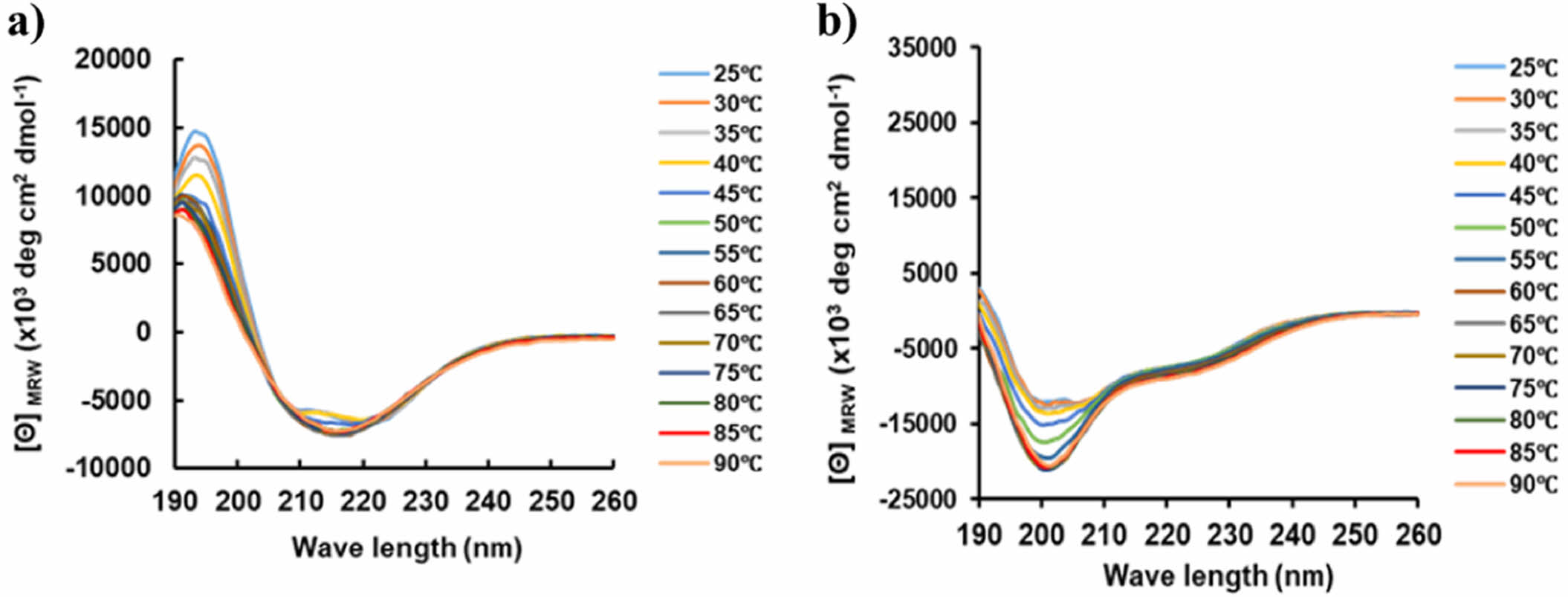
PrlF Antitoxin and YhaV Toxin Have Different Thermal Physiological Features at Varied Temperature conditions. In this study, the purified PrlF antitoxin and YhaV toxin were obtained as reported previously.9,18 As shown in Figure 2, the two proteins of PrlF and YhaV were well purified by Ni-NTA, and the molecular weights of each were confirmed to be 14386.1 Da and 19714.3 Da, respectively. Thermal-stability of TA systems are important for effective activity of the same and maintain its viability under high temperature conditions for suspension of growth under heat stress.22 The intrinsic thermal-stability of YhaV toxin was observed to be higher compared to its cognate PrlF antitoxin counterpart from 25 to 90 ℃ in Figure 3. The higher thermal-stability of the toxin can be the potential reason behind its activity under elevated temperature conditions responsible for arresting bacterial growth.23,24
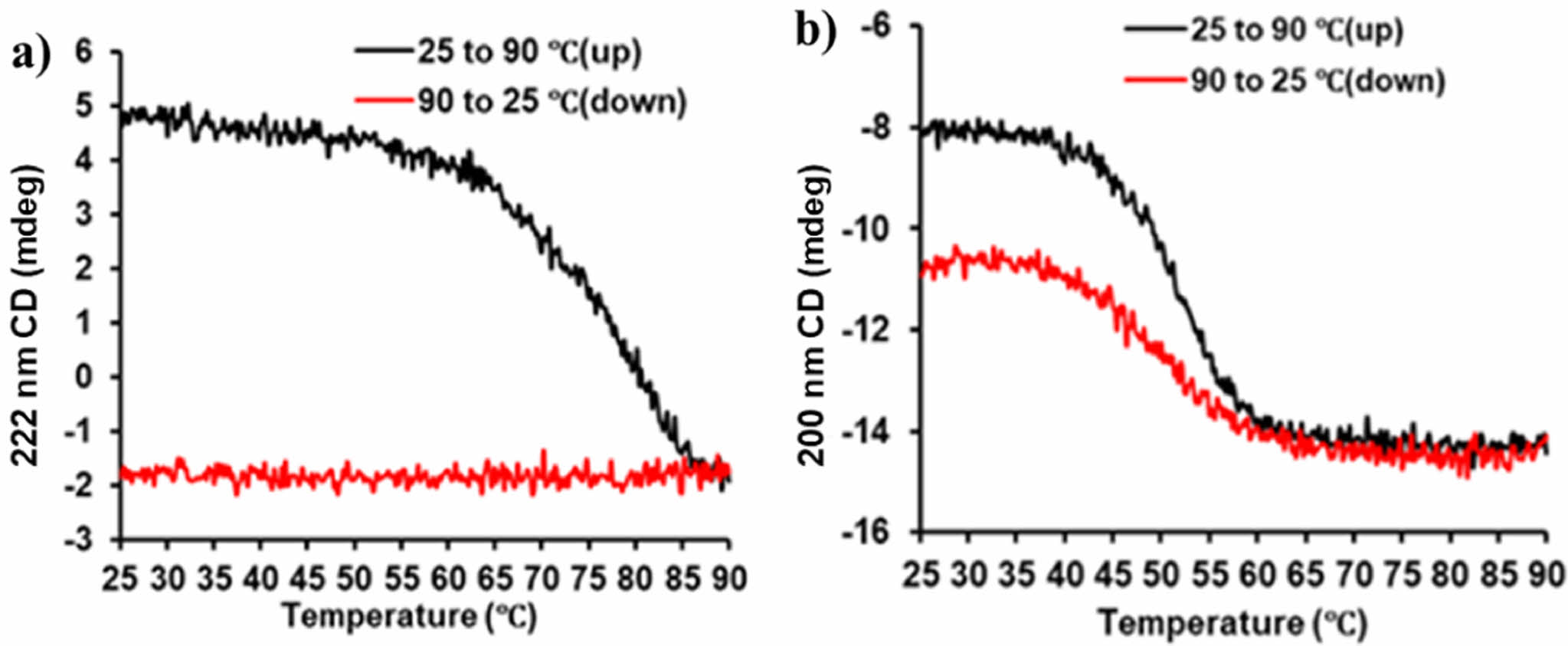
A previous study has also observed that the transcript levels of YhaV toxin was up-regulated under heat shock whereas no variation in transcript levels were observed for PrlF antitoxin.6 These features are characteristic for proteins belonging to the RelE superfamily.25 However, As shown in Figure 4. these melting points were recorded to be 70 ℃ for YhaV toxin and 50 ℃ for PrlF antitoxin. Furthermore, the CD spectrum of both the proteins at two different wavelengths, viz., 200 nm and 222 nm revealed the inability of YhaV toxin to refold into its native secondary structural conformation when the temperature was decreased from 90 to 25 ℃. On the other hand, the PrlF antitoxin was able to refold into the similar but not accurate structural conformation upon decrease in temperature. The CD spectrum at 25 to 90 ℃ temperature revealed that the antiparallel motif of the b-sheet region in PrlF provides flexibility, whereas the secondary motifs of YhaV was observed to be highly rigid one in Figure 5.
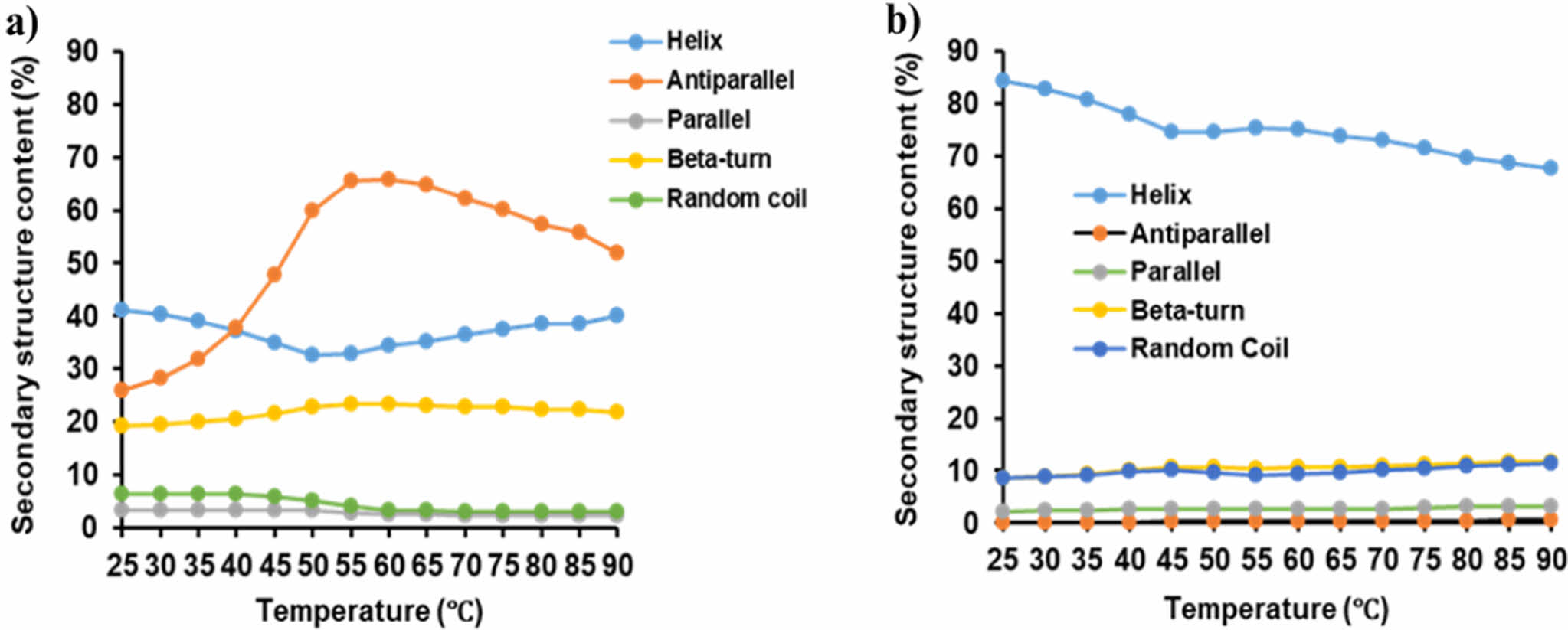
PrlF Antitoxin and YhaV Toxin Exhibit Distinct Secondary Structure. The secondary structural conformation is an important aspect for determining the functional characteristics of a particular protein.16,17 The principal component analysis (PCA) method based on the amino acid sequence that constitutes the protein is useful for analyzing the flexibility of protein structure, but it is difficult to analyze accurate folding and unfolding. In general, the structure of proteins is centered on hydrogen bonds, and in the case of a-helix proteins, bending and twisting are possible, but in the case of b-sheets, structural rigidity is usually present.28,29 As shown in Figure 5, the secondary component flexibility at 25 to 90 ℃ temperature revealed that the antiparallel motif of the β-sheet region in PrlF antitoxin provides flexibility, whereas the secondary motifs of YhaV toxin was observed to be highly rigid one.
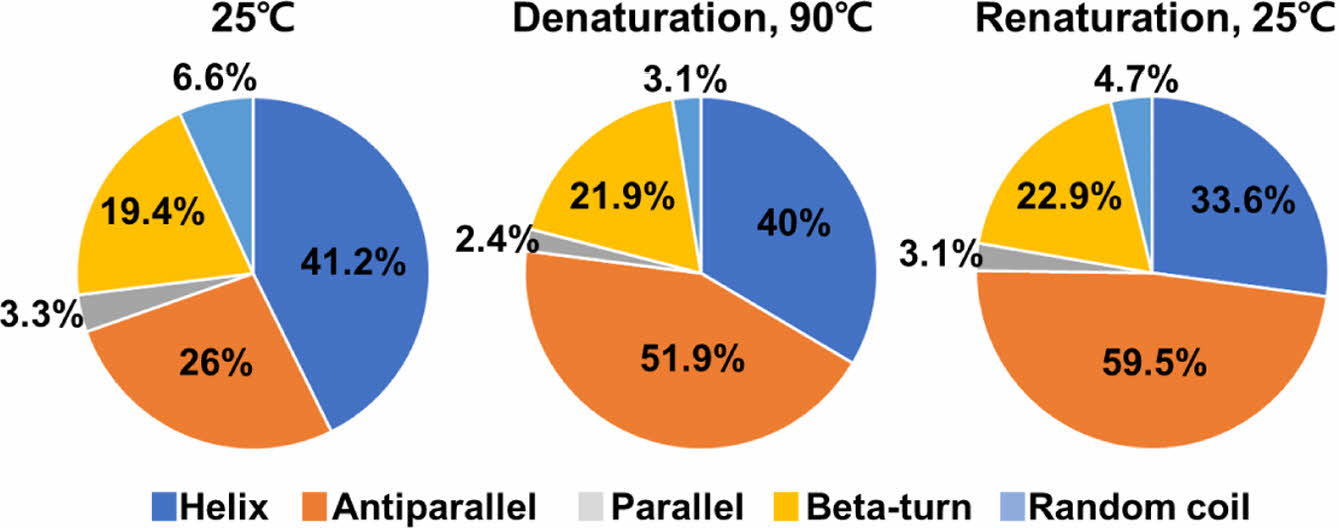
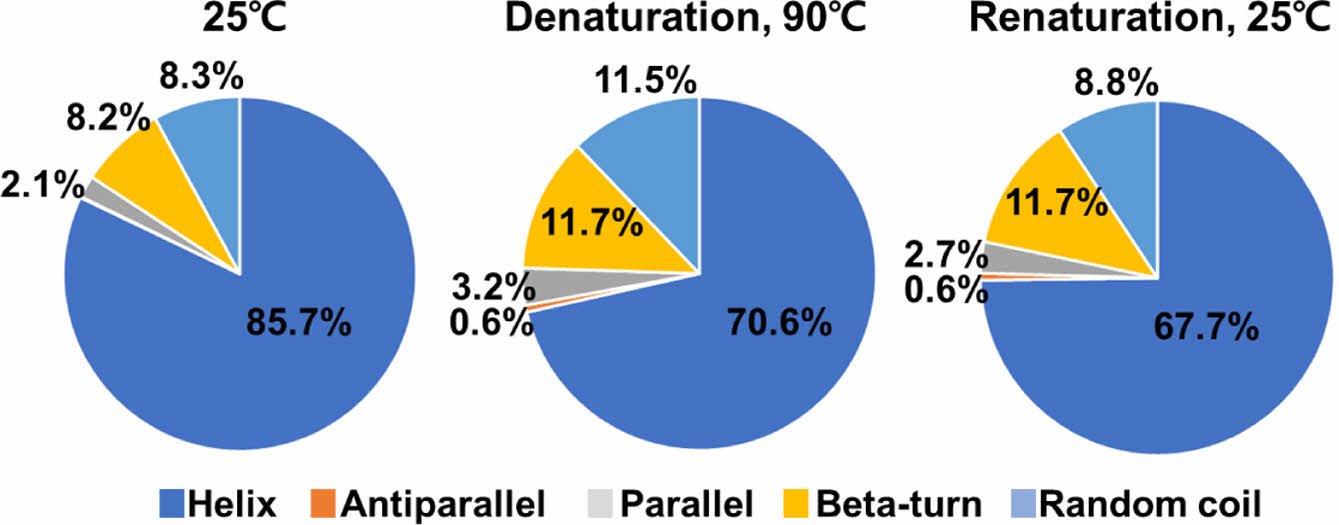
We compared changes in the secondary structure components of the two proteins under denaturation and renaturation conditions. The PrlF antitoxin was exhibited structural instability in both the helix and antiparallel components during the denaturation and renaturation states. Specially, the helix showed changes of 41.2%, 40%, and 33.6%, while the antiparallel exhibited changes of 26%, 51.9%, and 59.5% in its secondary structure composition in Figure 6. The YhaV toxin was found to exhibit structural stability in the helix during the denaturation and renaturation states, with changes in secondary structure content measured 85.7%, 70.6%, and 67.7% in Figure 7. Alpha helical proteins form a stable structure with a coiled coil form through C=O and N-H bonds of peptide bonds based on hydrogen bonds. This structure has structural stability because two alpha helices can twist each other with most of the nonpolar side chains on one side and these side chains face inward.
The higher number of α-helical motifs can be attributed to increase in hydrogen bonds which can result in providing thermal stability to YhaV toxin and function efficiently under high temperature conditions, whereas higher number of b-sheet motifs can result in lower thermal stability of PrlF antitoxin.26,27
|
Figure 1 Cell viability analysis using E. coli cells by FACS. Diagram of FACS results of (a) PrlF antitoxin, YhaV toxin, and PrlF-YhaV TA complex; (b) calculation percentages of cell viability. |
|
Figure 2 SDS-PAGE analysis of (a) purified PrlF and YhaV. Determination of (b) PrlF and YhaV molecular weights by MALDI-TOF. |
|
Figure 3 CD spectra showing the thermal flexibility of secondary structures of (a) YhaV; (b) PrlF over the temperature range of 25 to 90 ℃. |
|
Figure 4 Melting points and secondary structure reversibility of (a) YhaV; (b) PrlF. |
|
Figure 5 Secondary structure component flexibility of (a) PrlF; (b) YhaV from 25 to 90 ℃ |
|
Figure 6 Calculation of secondary structure component flexibilities of PrlF. |
|
Figure 7 Calculation of secondary structure component flexibilities of YhaV. |
In
conclusion, this study reveals the physiological importance of PrlF antitoxin in
inhibiting the bacteriostatic characteristic of YhaV toxin in vivo under
favourable growth conditions. The YhaV toxin consists of higher α-helix
residues in its secondary structure along with higher melting point, resulting
in enhanced
hydrogen bonding which can provide thermal stability to function under high
temperature conditions. The PrlF antitoxin had shown lower thermal stability
and its ability to refold into native secondary structural conformation during
decrease in temperature, that is important for functioning under normal environmental
conditions to neutralize the effect of toxin. Overall, this study puts
forward the important secondary structural conformation of PrlF antitoxin and
YhaV toxin in vitro for their efficient physiological functioning in Escherichia
coli.
- 1. Harms, A.; Broderson, D. E.; Mitarai, N.; Gerdes, K. Toxins, Targets, and Triggers: An overview of Toxin-Antitoxin Biology. Mol Cell. 2018, 70, 768-784.
-

- 2. Song, S.; Wood, T. K. Toxin-antitoxsin System Paradigms. Adv. Biosyst. 2020, 4, e1900290.
- 3. Schmidt, O.; Schuenemann, V. J.; Hand, N. J.; Silhavy, T. J.; Martin, J.; Lupas, A. N. PrlF and YhaV Encode a New Toxin-antitoxin System in Escherichia coli. J. Mol. Biol. 2007, 372, 894-905.
-

- 4. Habib, G.; Zhu, J.; Sun, B. A Novel Type I Toxin-antitoxin System Modulates Persister Cell Formation in Staphylococcus Aureus. IJMM. 2020, 310, 151400.
-

- 5. Frakin, N.; Goormaghtigh, F.; Van Melderne, L. Type II Toxin-Antitoxin Systems: Evoluation and Revolution. J. Bacteriol. 2020, 202, DOI:10.1128/jb.000763-19.
- 6. Muthuramalingam, M.; White, J. C.; Bourne, C. R. Toxin-antitoxin Modules are Pliable Switches Activated by Multiple Protease Pathway. Toxins. 2016, 8, 214.
-

- 7. Buts, L.; Lah, J.: Dao-Thi, M. H.; Wyns, L.; Loris, R. Toxin-antitoxin Modules as Bacterial Metabolisms as Bacterial Metabolic Stress Managers. Trends in Biochemical Sci. 2005, 30, 672-679.
-

- 8. Brzozowska, I.; Zielenkiewicz, U. Regulation of Toxin-antitoxin Systems by Proteolysis. Plasmid. 2013, 70, 33-41.
-

- 9. Choi, W.; Yamaguchi, Y.; Lee, J. W.; Jang, K. M.; Inouye, M.; Kim, S. G.; Yoon, M. H.; Park, J. H. Translation-dependent mRNA Cleavage by YhaV in Escherichia coli. FEBS let. 2017, 591, 1853-1861.
-

- 10. Han, Y.; Lee, E. J; Substrate Specificity of Bacterial Endoribonuclease Toxins. BMB Reports. 2020, 53, 611-621.
-

- 11. Gotfredsen, M.; Gerdes, K. The Escherichia coli relBE Genes Belong to a New Toxin-antitoxin Gene Family. Mol. Microbiol. 1998, 29, 1065-1076.
-

- 12. Overgaard, M.; Borch, J.; Jorgensen, M. G.; Gerdes, K. Messenger RNA Interferase RelE Controls relBE Transcription by Conditional Cooperativity. Mol. Microbial. 2008, 69, 841-857.
-

- 13. Wang, Y.; Wang, H.; Hay, A. J.; Zhong, Z.; Zhu, J.; Kan, B. Functional RelBE Family Toxin-antitoxin Pairs Affect Biofilm Maturation and Intestine Colonization in Vibrio Cholerae. Plos One. 2015, 10, e0135696.
-

- 14. Neubauer, C.; Gao, Y. G.; Andersen, K. R.; Dunham, C. M.; Kelley, A. C.; Hentschel, J. The Structural Basis for mRNA Recognition and Cleavage by the Ribosome-dependent Endonuclease RelE. Cell. 2009, 139, 1084-1095.
-

- 15. Choi, W.; Yoon, M. H.; Park, J. H. Functional Characterization of the C-terminus of YhaV in the Escherichia coli PrlF-YhaV Toxin-antitoxin System. J. Microbial. Biotechnol. 2018, 28, 987-996.
-

- 16. King, R. D.; Sternberg, M. J. Identification and Application of the Concepts Important for Accurate and Reliable Protein Secondary Structure Prediction. Protein Sci. 1996, 5, 2298-2310.
-

- 17. Baldi, P.; Brunak, S.; Frasconi, P.; Soda, G.; Pollastri, G. Exploiting the Past and the Future in Protein Secondary Structure Prediction. Bioinformatics. 1998, 15, 937-946.
-

- 18. Nebe Von Caron, G.; Stephens, P. J.; Hewitt, C. J.; Powell, J. R.; Badley, R. A. Analysis of Bacterial Function by Multi-color Fluorescence Flow Cytometry and Single Cell Sorting. J. Microbial. Methods. 2000, 42, 97-1147.
-

- 19. Kim, S. G.; Shin, S. Y.; Park, Y. C.; Shin, C. S.; Seo, J. H. Production and Solid-phase Refolding of Human Glucagon-like Peptide-1 Using Recombinant Escherichia coli. Protein Expression and Purification. 2011, 79, 197-203.
-

- 20. Lee, Y. G.; Lee, S. H.; Chung, Y. H.; Kim, S. I. Characterization of Thermostable Deblocking Aminopeptidases of Archaeon Thermococcus Onnurineus NA1 by Proteomic and Biochemical Approaches. J. Microbiol. 2012, 50, 792-797.
-

- 21. Bohm, G.; Muhr, R.; Jaenicke, R. Quantitative Analysis of Protein Far-UV Circular Dichroism Spectra by Neural Networks. Protein Engineering. 1992, 5, 191-195.
-

- 22. Cherny, I.; Overgaard, M.; Borch, J.; Bram, Y.; Gerdes, K.; Gazit, E. Structural and Thermodynamic Characterization of the Escherichia coli RelBE Toxin-antitoxin System: Indication for a Functional Role of Differential Stability. Biochemistry. 2007, 46, 12152-12163.
-

- 23. Yamaguchi, Y.; Inouye, M. mRNA Interferase, Sequence-specific Endoribonuclease From the Toxin-antitoxin System. Progress in Molecular Biology and Translational Science. 2009, 85, 467-500.
-

- 24. Yamaguchi, Y.; Inouye, M. Regulation of Growth and Death in Escherichia coli by Toxin-antitoxin System. Nat. Rev. Microbiol. 2011, 9, 779-790.
-

- 25. Francuski, D.; Saenger, W. Crystal Structure of the Antitoxin-toxin Protein Complex RelB-RelE From Methanococcus Jannaschii. J. Mol. Biol. 2009, 393, 898-908.
-

- 26. Scholtz, J. M.; Baldwin, R. L. The Mechanism of Alpha-helix Formation by Peptides. Annual Reviews of Biophysics and Biomolecular Structure. 1992, 21, 95-118.
-

- 27. Numata, K.; Cebe, P.; Kaplan, D. L. Mechanism of Enzymatic Degradation of Beta-sheet Crystal. Biomaterials. 2010, 31, 2926-2933.
-

- 28. Emberly, E. G.; Mukhopadhyay, R.; Wingreen, N. S.; Tang, C. Flexibility of Alpha-helices: Results of a Statistical Analysis of Database Protein Structure. J. Molecular Biology. 2003, 327, 229-237.
-

- 29. Numata, K.; Cebe, P.; Kaplan, D. L. Flexibitliy of Neta-sheets: Principal Component Analysis of Database Protein Structures. Proteins. 2004, 55, 91-98.
- Polymer(Korea) 폴리머
- Frequency : Bimonthly(odd)
ISSN 2234-8077(Online)
Abbr. Polym. Korea - 2024 Impact Factor : 0.6
- Indexed in SCIE
 This Article
This Article
-
2025; 49(6): 693-699
Published online Nov 25, 2025
- 10.7317/pk.2025.49.6.693
- Received on Jan 22, 2025
- Revised on Apr 15, 2025
- Accepted on Apr 22, 2025
 Services
Services
- Full Text PDF
- Abstract
- ToC
- Acknowledgements
- Conflict of Interest
Introduction
Experimental
Results and Discussion
Conclusions
- References
Shared
 Correspondence to
Correspondence to
- Yong-kyu Lee
-
*4D Convergence Technology Institute (National Key Technology Institute in University),
Korea National University of Transportation, Jungpyeog, Chung-Buk 27909, Korea
**Department of Chemical & Biological Engineering, Korea National University of Transportation,
Chungju, Chung-Buk 27469, Korea - E-mail: leeyk@ut.ac.kr









 Copyright(c) The Polymer Society of Korea. All right reserved.
Copyright(c) The Polymer Society of Korea. All right reserved.